
#Its sequence analysis pdf
Although some groups are under- or unrepresented in the two data sets due to, e.g. Analysis And Its Applications In Pdf can be taken as well as picked to act. To analyze the N-terminal amino acid sequence, the purified enzymes were blotted onto PVDF membranes (Bio-Rad Laboratories, CA, USA) and sequenced on an Applied Biosystems 477A-120A sequencer according to the manufacturers instructions (Applied Biosystemes, CA, USA).
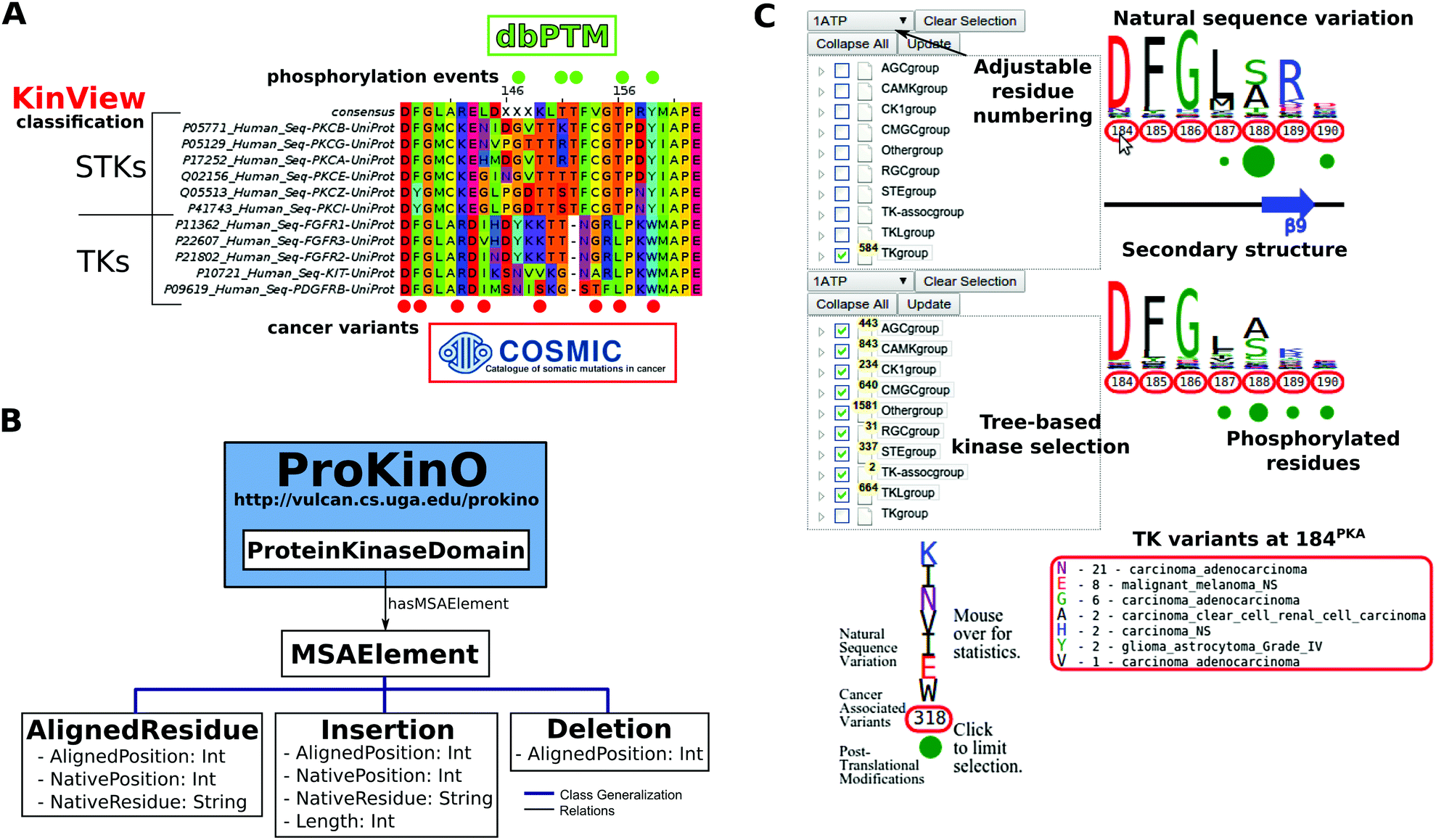
Since the 1990s, the Sanger dye-termination sequencing method has been the most widely used method of automated DNA sequencing of VP7 and VP4 RVA genes. Clustering analyses of the 134 692 ITS1 and ITS2 pyrosequences using a 97% similarity cut-off revealed a high similarity between the two data sets when it comes to taxonomic coverage. Sequence analysis is the definitive method for confirmation of RVA genotypes. However, no single threshold value worked well for all fungi at the same time within the curated UNITE database, and we found that the Operational Taxonomic Unit (OTU) concept is not easily translated into the level of species because many species are distributed over several clusters. Clustering analyses of sequences with known taxonomy using the bioinformatics pipeline ClustEx revealed that a 97% similarity cut-off represent a reasonable threshold for estimating the number of known species in the data sets for both ITS1 and ITS2. We analyse three data sets: two comprising ITS1 and ITS2 sequences of known taxonomic affiliations and a third comprising ITS1 and ITS2 environmental amplicon pyrosequencing data. ITS2 as a DNA metabarcoding marker for fungi.

In event sequences, positions do not convey any time information. This could be the historical calendar time, or a process time such as age, i.e. In this methodological study we evaluate the usability of ITS1 vs. RDP provides quality-controlled, aligned and annotated Bacterial and Archaeal 16S rRNA sequences, and Fungal 28S rRNA sequences, and a suite of analysis. Basic concept of sequence analysis in social sciences A crucial point when looking at state sequences is the timing scheme used to time align the sequences. In amplicon pyrosequencing studies of fungal diversity, one of the spacers ITS1 or ITS2 of the ITS region is normally used. The approach is frequently used in population and community microbial ecology studies, phylogenetic reconstruction of target microbial groups, identification of individual species in pure cultures, and detection of organisms of interest (pathogens or beneficial), among many others.The nuclear ribosomal Internal Transcribed Spacer ITS region is widely used as a DNA metabarcoding marker to characterize the diversity and composition of fungal communities. Novogene amplicon metagenomic sequencing services provide to efficiently screen for variants and target organisms, and describe and compare the diversity of multiple complex environments. Amplicon metagenomic sequencing is designated to sequence the target genes of 16S ribosomal RNA (rRNA), or 18S rRNA and Internal Transcribed Spacer (ITS) rRNA by universal primers, to describe and compare the phylogeny and taxonomy of bacteria (and archaea) and fungi (such as yeasts, molds and etc.), respectively. Short (<500 bp) hypervariable regions of conserved genes or intergenic regions are amplified by PCR and analyzed by next generation sequencing (NGS) technology, to identify and differentiate multiple microbial species from complicated samples. Sequence analysis: Its past, present, and future Authors: Tim Futing Liao University of Illinois, Urbana-Champaign Danilo Bolano Universit commerciale Luigi Bocconi Christian Brzinsky-Fay. 16S/18S/ITS Amplicon Metagenomic Sequencing is an ultra-deep DNA sequencing method that focuses on sequencing specific target regions (amplicons).


 0 kommentar(er)
0 kommentar(er)
